mqsA
antitoxin for MqsR toxin; transcriptionalrepressorBBF10K_001108
source
Escherichia coli str. K-12 substr. MG1655
Antitoxin component of a type II toxin-antitoxin (TA) system. Labile antitoxin that binds to the MqsR mRNA interferase toxin and neutralizes its endoribonuclease activity. Overexpression prevents MqsR-mediated cessation of cell growth and inhibition of cell proliferation. Initially reported to act as a cotranscription factor with MqsA (PubMed:19690171, PubMed:20105222). Following further experiments, the MqsR-MqsA complex does not bind DNA and all reported data are actually due to a small fraction of free MqsA alone binding DNA. Addition of MqsR to a preformed MqsA-promoter DNA complex causes dissociation of the MqsA-DNA complex, probably causing derepression of MqsA-repressed transcripts (PubMed:23172222). MqsA binds to 2 palindromes in the promoter region of the mqsRA operon activating its transcription. Binds to other promoters, inducing mcbR and spy and repressing cspD among others (PubMed:20105222). Binds to and represses the rpoS promoter, the master stress regulator, resulting in decreased cyclic-di-GMP, reduced stress resistance, increased cell motility and decreased biofilm formation; in these experiments 5 TA systems are missing (lacks MazEF, RelEB, ChpB, YoeB-YefM, YafQ-DinJ) (PubMed:21516113). An earlier study showed overexpression alone increases biofilm formation, perhaps by repressing cspD; in these experiments the 5 TA systems are present (PubMed:20105222). Represses the csgD promoter. In the presence of stress, when this protein is degraded, the promoters it represses are derepressed, leading to biofilm formation (Probable). This TA system mediates cell growth during bile acid deoxycholate stress by degrading mRNA for probable deoxycholate-binding protein YgiS; bile acid detergents such as deoxycholate are important for host defense against bacterial growth in the gall bladder and duodenum (PubMed:25534751).
attr.
Keoni Gandall
Usage
growth
shipping strain
{shipping_strain}
growth conditions
37 C, shaking 300 rpm
antibiotic
ampicillin
expression
strain
N/A
promoter
N/A
inducer
N/A
cloning
method
GoldenGate
enzyme
BsaI
overhangs
3' - AATG … GCTT - 5'
sequencing
forward primer
M13 For
reverse primer
M13 Rev
Construct
plasmid name
pOpen-mqsA
plasmid size
2519
insert size
396
origin
ColE1 High Copy
copy number
500-700
Safety
BSL
BSL1
other information
No Value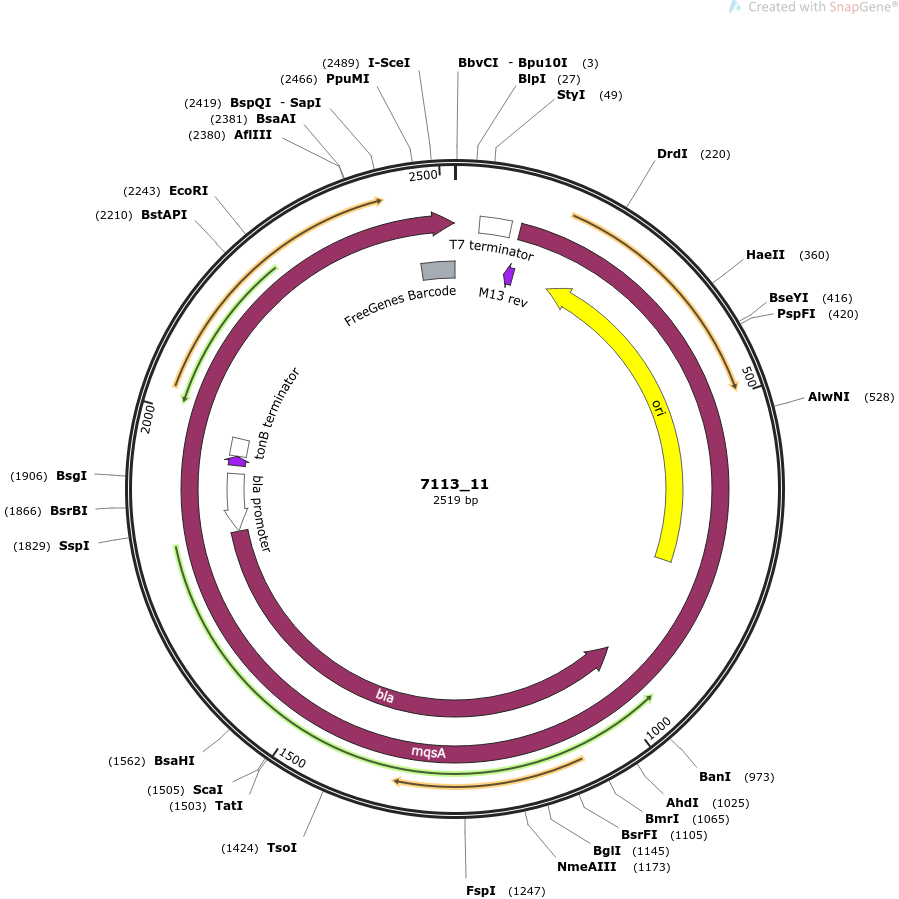
References
Available Elsewhere
FALSE