gyrB
DNA gyrase subunit BBBF10K_001188
source
Escherichia coli str. K-12 substr. MG1655
DNA gyrase negatively supercoils closed circular double-stranded DNA in an ATP-dependent manner to maintain chromosomes in an underwound state (PubMed:186775, PubMed:3031051, PubMed:1323022, PubMed:8248233, PubMed:7811004, PubMed:8621650, PubMed:9657678, PubMed:12051842, PubMed:12051843, PubMed:18642932, PubMed:19060136, PubMed:19965760, PubMed:22457353, PubMed:23294697, PubMed:20356737, PubMed:20675723, PubMed:23352267, PubMed:24386374, PubMed:25202966, PubMed:25849408). This makes better substrates for topoisomerase 4 (ParC and ParE) which is the main enzyme that unlinks newly replicated chromosomes in E.coli (PubMed:9334322). Gyrase catalyzes the interconversion of other topological isomers of double-stranded DNA rings, including catenanes (PubMed:22457352). Relaxes negatively supercoiled DNA in an ATP-independent manner (PubMed:337300). E.coli gyrase has higher supercoiling activity than other characterized bacterial gyrases; at comparable concentrations E.coli gyrase introduces more supercoils faster than M.tuberculosis gyrase, while M.tuberculosis gyrase has higher decatenation than supercoiling activity compared to E.coli (PubMed:22457352). E.coli makes 15% more negative supercoils in pBR322 plasmid DNA than S.typhimurium; the S.typhimurium GyrB subunit is toxic in E.coli, while the E.coli copy can be expressed in S.typhimurium even though the 2 subunits have 777/804 residues identical (PubMed:17400739). The enzymatic differences between E.coli gyrase and topoisomerase IV are largely due to the GyrA C-terminal domain (approximately residues 524-841) and specifically the GyrA-box (PubMed:8962066, PubMed:16332690).
attr.
Keoni Gandall
Usage
growth
shipping strain
{shipping_strain}
growth conditions
37 C, shaking 300 rpm
antibiotic
ampicillin
expression
strain
N/A
promoter
N/A
inducer
N/A
cloning
method
GoldenGate
enzyme
BsaI
overhangs
3' - AATG … GCTT - 5'
sequencing
forward primer
M13 For
reverse primer
M13 Rev
Construct
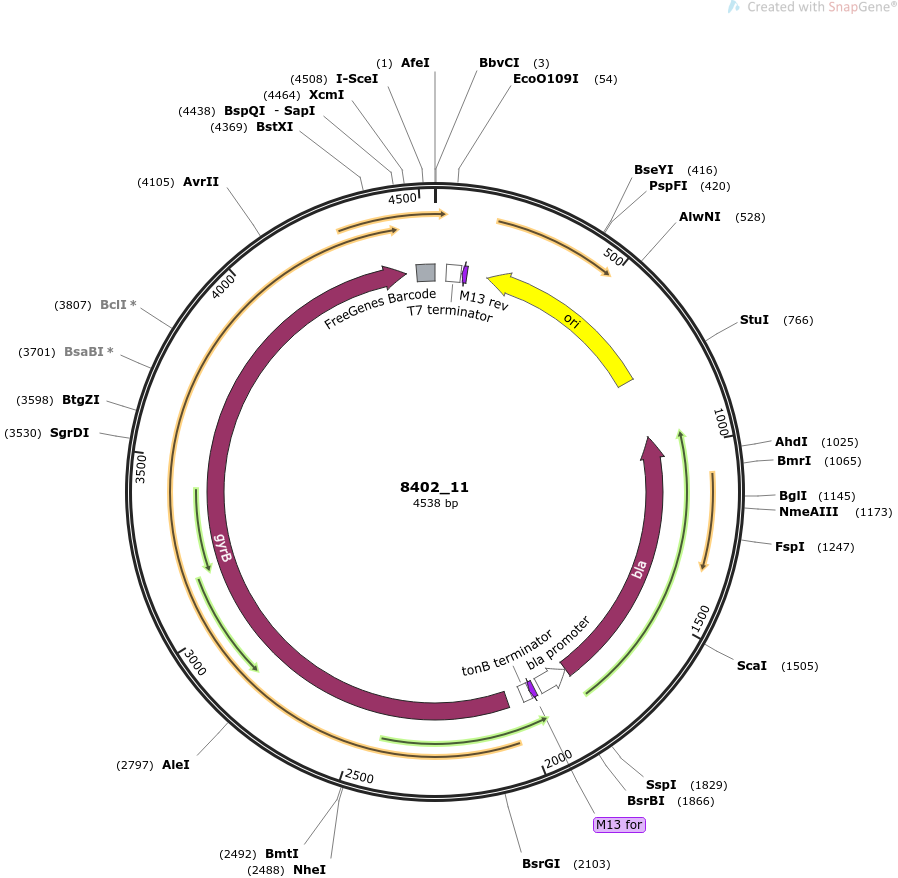
plasmid name
pOpen-gyrB
plasmid size
4538
insert size
2415
origin
ColE1 High Copy
copy number
500-700
Safety
BSL
BSL1
other information
No ValueReferences
Available Elsewhere
FALSE