K12polLF
DNA Polymerase I, Large (Klenow) FragmentBBF10K_003248
source
Escherichia coli (strain K12)
DNA polymerase fragment that retains 5' → 3' polymerase activity and 3’ → 5’ exonuclease activity for removal of precoding nucleotides and proofreading, but loses 5' → 3' exonuclease activity. Usable for synthesis of double-stranded DNA from single-stranded templates, filling in of receded 3' ends of DNA fragments to make 5' overhang blunt, digesting away protruding 3' overhangs, preparation of radioactive DNA probes.
attr.
Chiara Gandini, Open Bioeconomy Lab
Usage
growth
shipping strain
{shipping_strain}
growth conditions
37 C, shaking 300 rpm
antibiotic
ampicillin
expression
strain
N/A
promoter
N/A
inducer
N/A
cloning
method
GoldenGate
enzyme
BsaI
overhangs
3' - AATG … GCTT - 5'
sequencing
forward primer
M13 For
reverse primer
M13 Rev
Construct
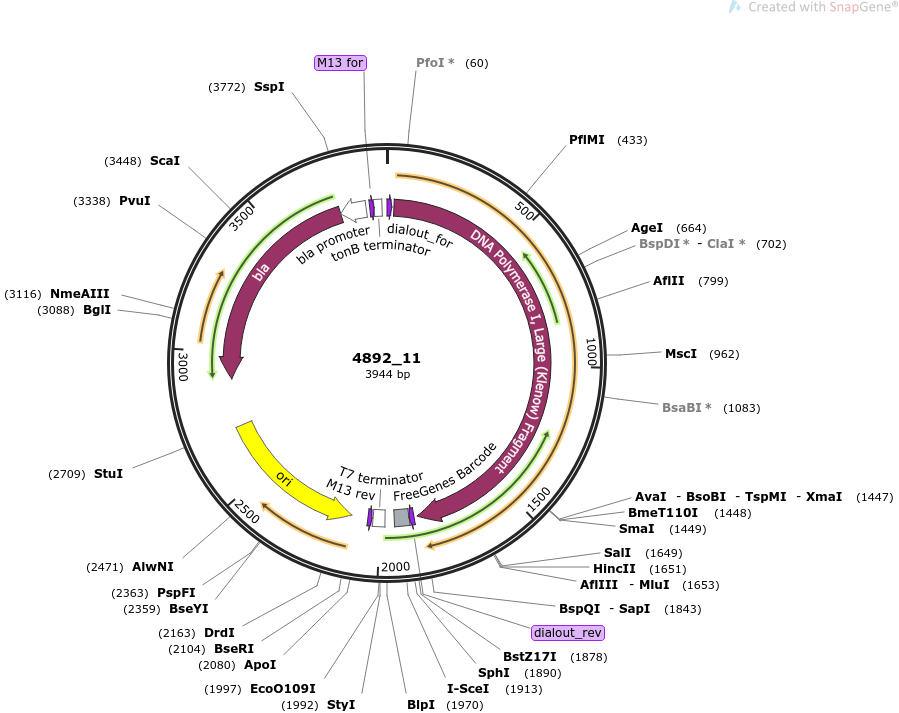
plasmid name
pOpen-K12polLF
plasmid size
3944
insert size
1821
origin
ColE1 High Copy
copy number
high (3-500)
Safety
BSL
BSL1
other information
No ValueReferences
Available Elsewhere
FALSE